Scientific Scope
Whole genome sequencing has provided new insights into the fundamentals of health and disease. This information forms the basis for the development of new diagnoses and ultimately new therapies. Our laboratory uses various strategies to decipher gene function and employ gene editing tools to discover new disease markers and develop innovative cure strategies
Research Areas

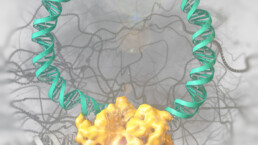
The growing number of published genome sequences has highlighted the need for precise and efficient genomic manipulation strategies. Genetic engineering tools, such as site-specific recombinases, allow for the predictable and precise manipulation of whole organisms. To enhance the capabilities of these enzymes, we are utilizing a combination of directed molecular evolution, rational design, and AI to develop novel recombinases. Example are the development of a recombinase, which can specifically remove the HIV-1 provirus from infected cells and a recombinase system, that can correct a large genomic inversion found in severe Hemophilia A patients. In future research, we aim to expand the use of designer-recombinases for therapeutic applications.

Programmable nucleases offer a powerful tool for targeting mutations in cancer cells. However, nucleases depend on cellular DNA repair pathways that are hard to control and can lead to unfavorable editing. In contrast, base and prime editors can correct mutations in both oncogenes and tumor suppressor genes at high precision, highlighting their potential for precision oncology. Our goal is to develop these systems for identifying patient-specific vulnerabilities and as a new therapeutic modality.

Epigenome engineering offers the capability to change epigenetic marks at specific genomic locations to control gene expression without altering the genetic code. We are researching novel epigenetic editors that can effectively silence genes, regardless of their genomic context or cell type. Results from these experiments should improve our knowledge about the epigenome and built the foundation for novel therapeutic modalities.
Future prospects and goals
- Improve esiRNA and CRISPR/Cas9 technology for functional genomic studies.
- Perform phenotypic profiling of cancer cell lines and primary cancer material to identify and characterize candidate genes with therapeutic potential.
- Develop and apply tools that allow flexible and precise genomic manipulations.